QIASURE METHYLATION TEST CAN DECREASE CLINICALLY NON RELEVANT COLPOSCOPY REFERRALS OF HPV POSITIVE WOMEN WITH ASC-US/ LSIL CYTOLOGY.
British Journal of Cancer; https://doi.org/10.1038/s41416-021-01614-4
Primary HPV screening has doubled the number of colposcopy referrals because of direct referral of HPV-positive women with ASC-US/ LSIL cytology.
This study evaluated the CIN3+ risks and colposcopy referrals by
– QIAsure Methylation Test analysis
– HPV16/18 genotyping
– HPV16/18/31/33/45 genotyping
in HPV-positive women with ASC-US/ LSIL (BMD) in two Dutch screening trials.
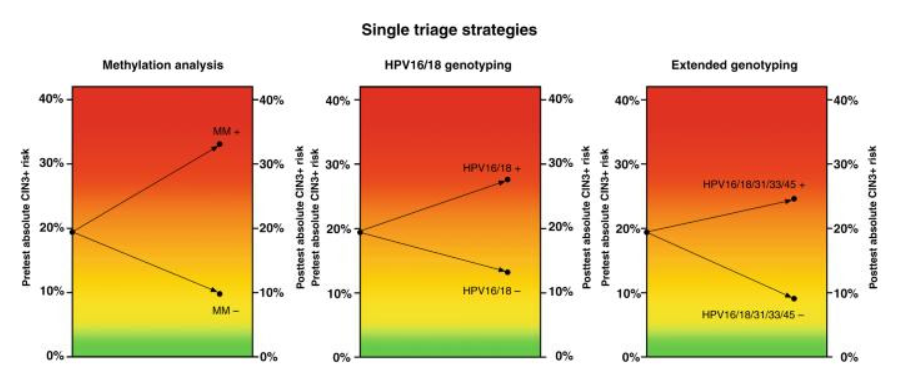
– QIAsure Methylation Test discriminated well, yielding a CIN3+ risk of 33.1% after a positive result and a CIN3+ risk of 9.8% after a negative result. Colposcopy referral percentages were 41.2% for QIAsure Methylation Test.
– HPV16/18 and HPV16/18/31/33/45 genotyping resulted in a 27.6% and 24.6% CIN3+ risk after a positive result, and a 13.2% and 9.1% CIN3+ risk after a negative result, discriminating less than QIAsure Methylation Test. Colposcopy referral percentages were 43.2%, and 66.3% for HPV16/18 and HPV16/18/31/33/45 genotyping, respectively.
CONCLUSION:
The use of QIAsure Methylation Test in HPV-positive women with ASC-US/ LSIL (BMD) cytology can lead to a substantial reduction in the number of direct colposcopy referrals.